Andrew White | FutureHouse
Automating Literature Research
FutureHouse
Health Research Alliance
March 2025
FutureHouse Structure
- Non-profit
- Funded primarily by Eric Schmidt
- Based in San Francisco
- 20 employees
Science is changing independent of AI
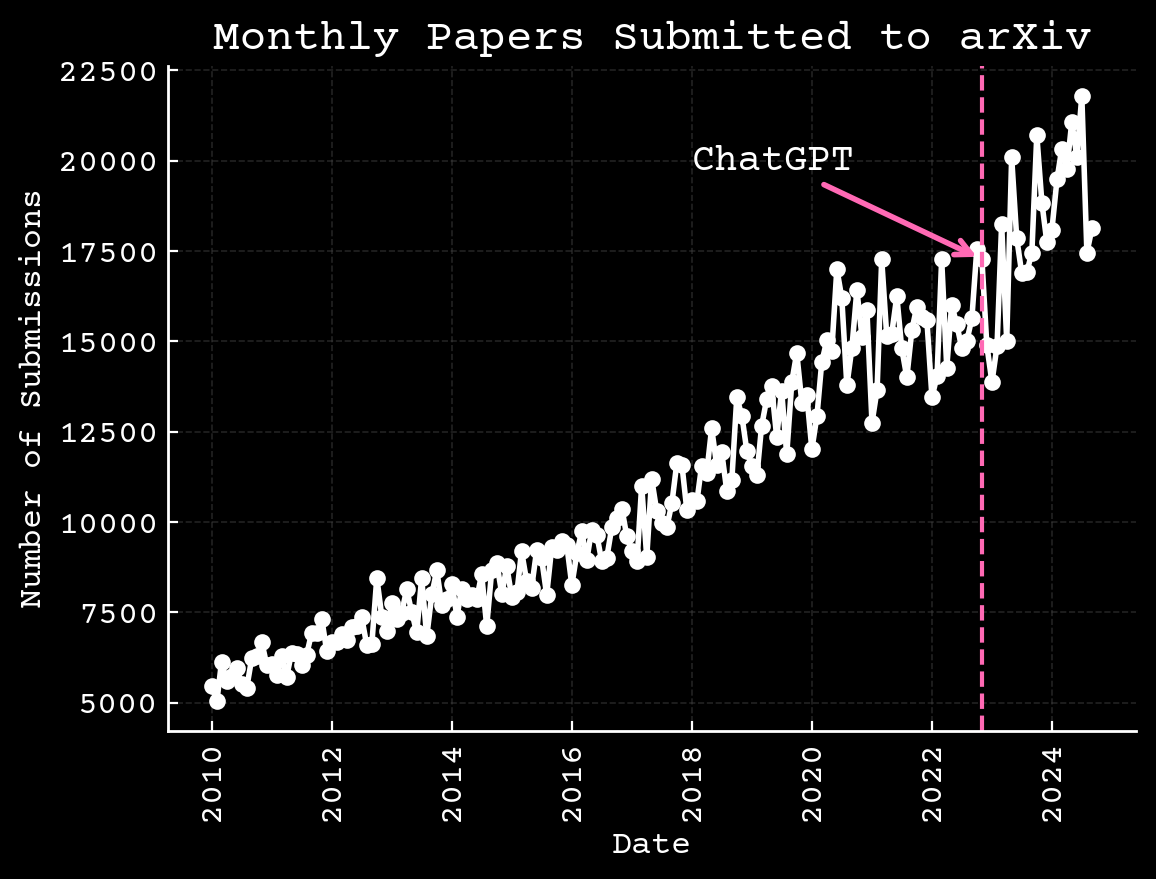
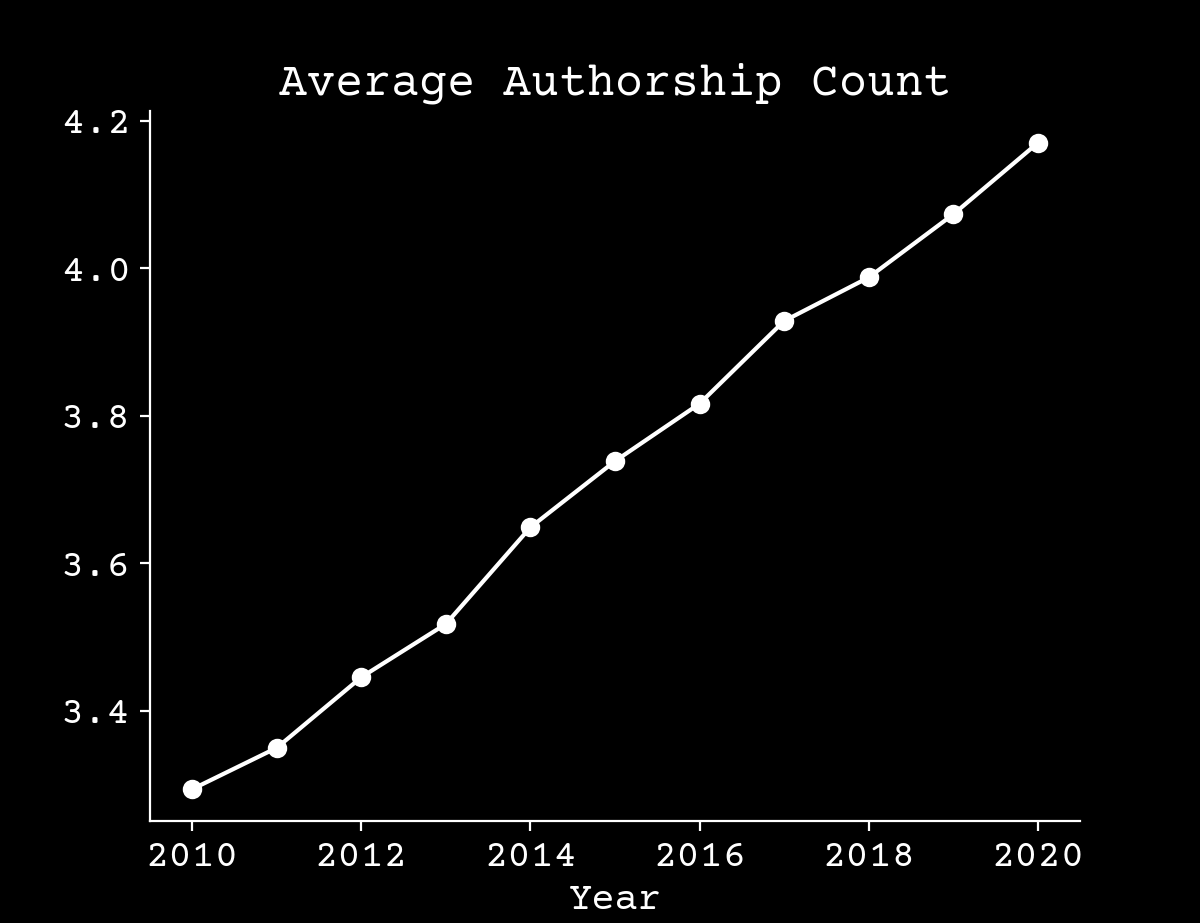
Arxiv.org,10.6084/m9.figshare.17064419.v3
Intellectual bottlenecks are growing
📝 Increasing paper count ($\approx$5M per year)
🧬 Larger data sets from cheaper experiments (genome at
$200 per person, $1 / GB of sequencing)
🔍Increasingly less disruptive papers (96% decline in biology)
Mission
Accelerate Scientific Discovery with Language Agents
Can LLMs do science already?
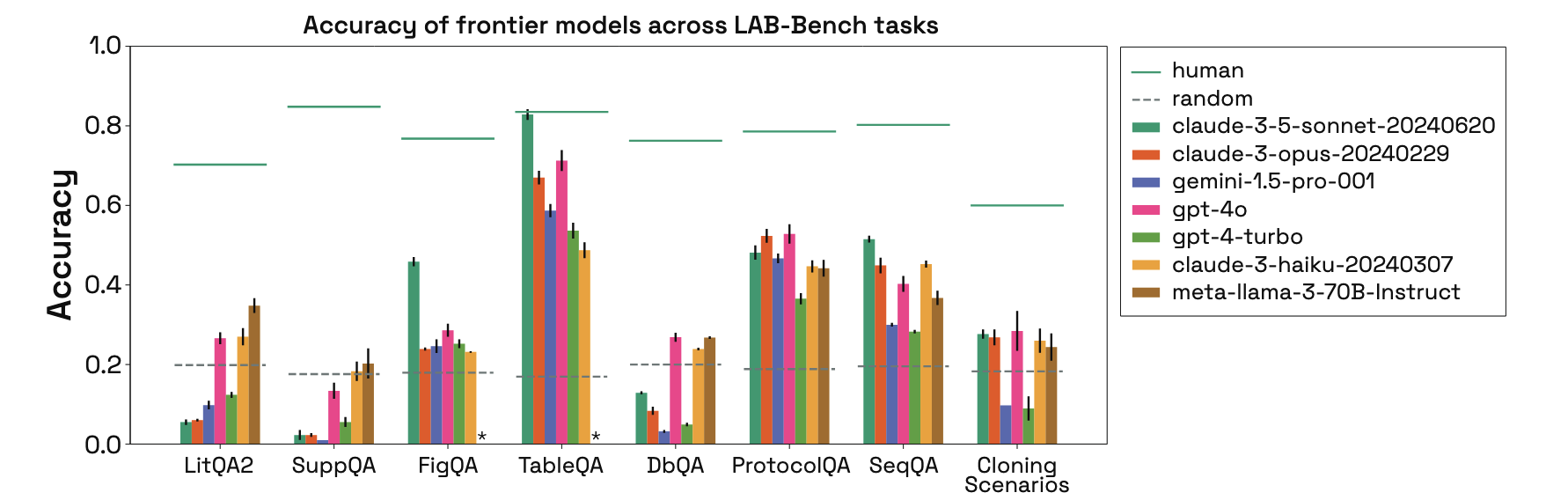
LAB-Bench: Measuring Capabilities of Language Models for Biology Research
Jon M. Laurent, Joseph D. Janizek, Michael Ruzo, Michaela M. Hinks, Michael J. Hammerling, Siddharth Narayanan, Manvitha Ponnapati, Andrew D. White, Samuel G. Rodriques arXiv:2407.10362, 2024
Existing benchmarks
MMLU-Pro
- As of 2017, how many of the world's 1-year-old children today have been vaccinated against some disease?
- Find the logarithm of 3^2
Lab-Bench Questions
Not textbook knowledge
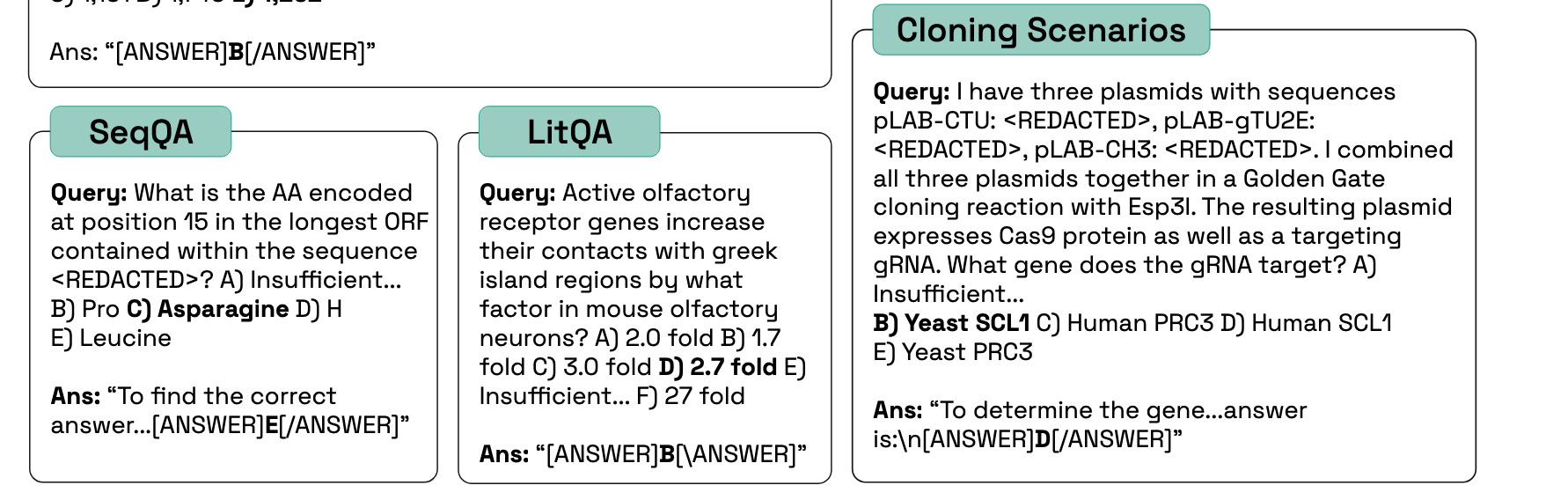
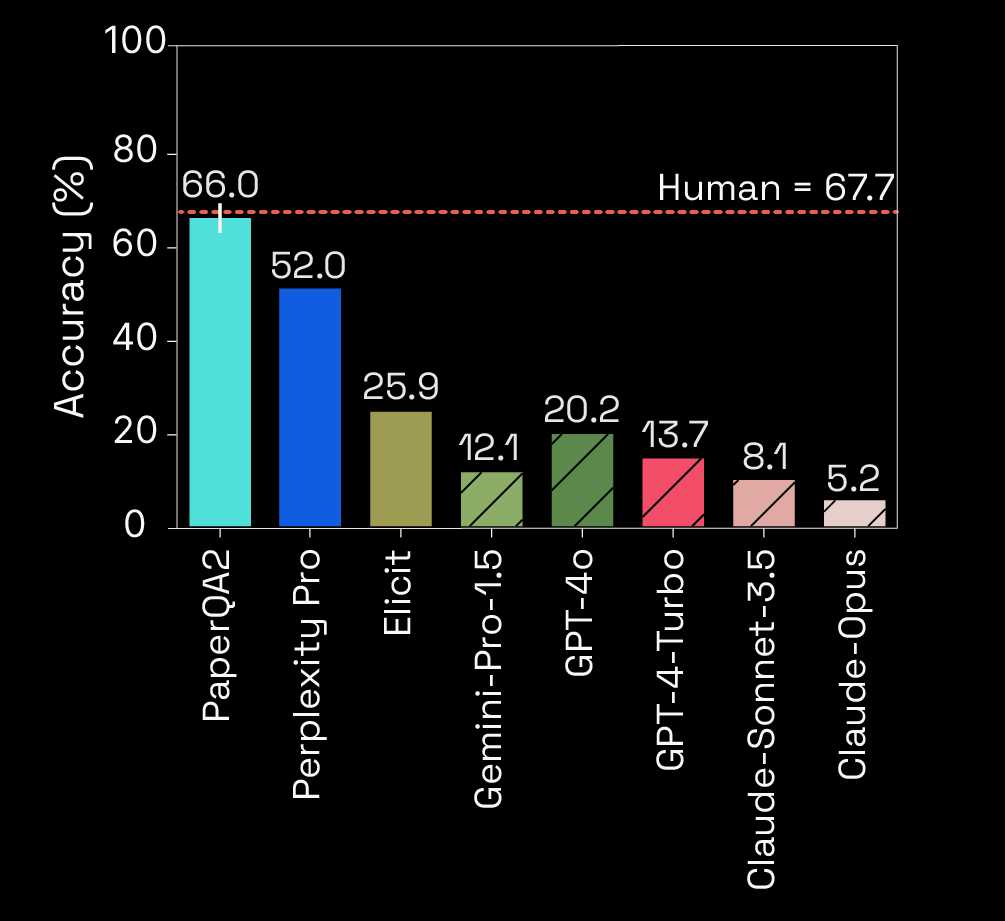
Human baselines exceed LLMs
LitQA2 Question
Which three residues with evolutionary divergence in the G domains of RAS isoforms also impose selectivity constraints of pan-KRAS non-covalent inhibition?
GPT-4 Response (prompted to cite)
The three residues with evolutionary divergence in the G domains of KRAS, HRAS, and NRAS isoforms that impose selectivity constraints for pan-KRAS non-covalent inhibition are Thr35, Asp38, and Glu37. It has been observed that these three amino acids significantly influence the binding affinity and selectivity of potential non-covalent inhibitors, thereby determining the efficacy of different therapeutic strategies targeting the RAS isoforms (Hunter et al., 2015). The understanding of these residues' evolutionary conservation and differences across isoforms can aid the development of more effective pan-RAS inhibitors (Lu et al., 2016).
Wrong, hallucinated references
PaperQA Agent Response
Three key residues in the G domains of RAS isoforms that affect KRAS inhibitor selectivity have been identified:
- H95 (in the α3 helix): In KRAS this position contains histidine (H), while it is leucine (L) in NRAS and glutamine (Q) in HRAS. H95 exerts orthosteric (direct) effects on inhibitor selectivity (kim2023pankrasinhibitordisables pages 3-4, parker2018thekrasnras pages 5-7).
- P121: This position contains proline (P) in both KRAS and NRAS, but alanine (A) in HRAS. P121 has allosteric (indirect) effects on selective inhibition (kim2023pankrasinhibitordisables pages 3-4, parker2017themostoncogenic pages 28-33).
- S122 (in loop 8): This position varies across all three isoforms, with serine (S) in KRAS, threonine (T) in NRAS, and alanine (A) in HRAS. Like P121, S122 contributes to selectivity through allosteric effects (kim2023pankrasinhibitordisables pages 3-4, parker2018thekrasnras pages 7-9).
What is an agent?
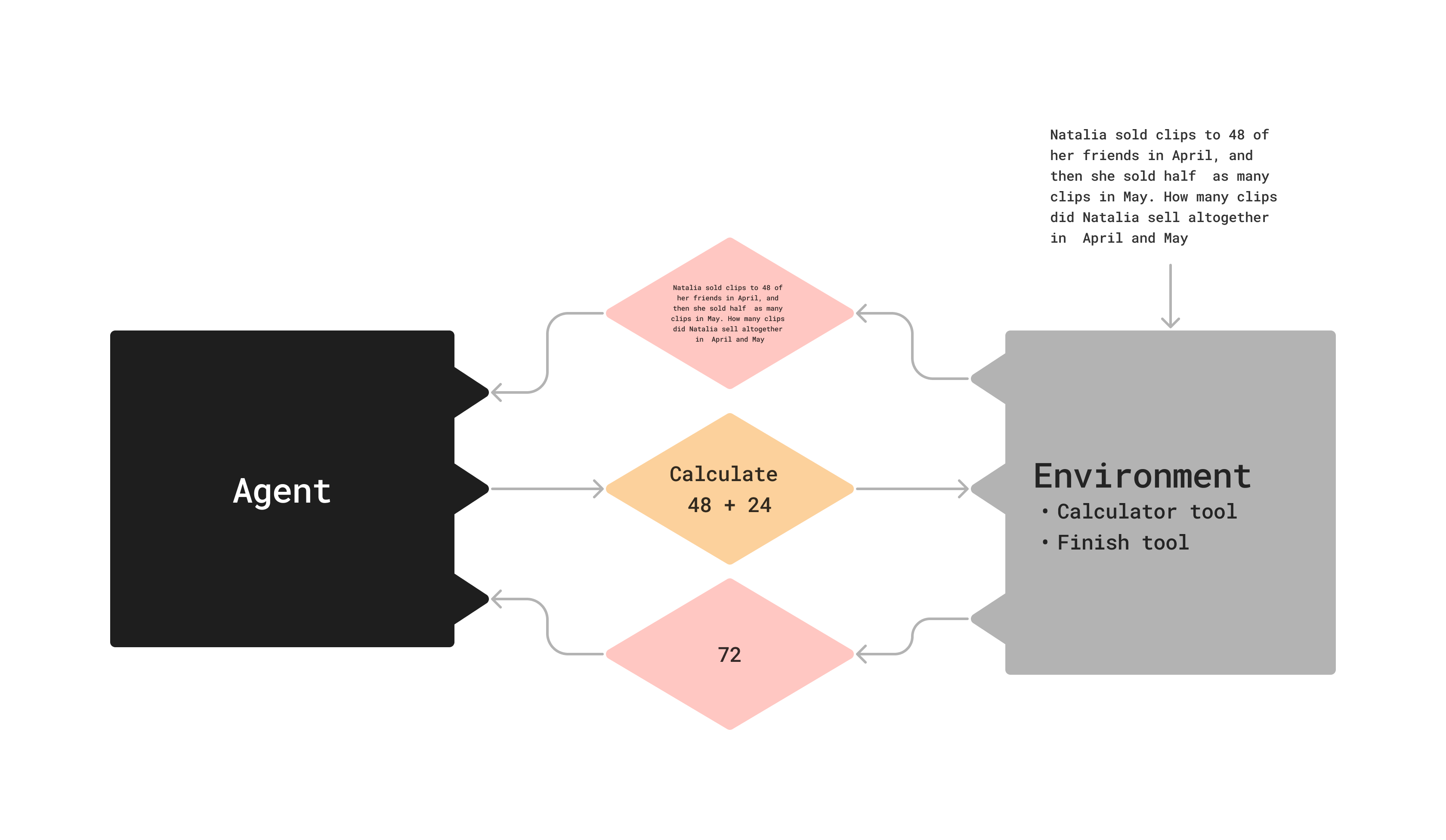
Agent: trained, makes decisions
Environment: untrained, has tools, state
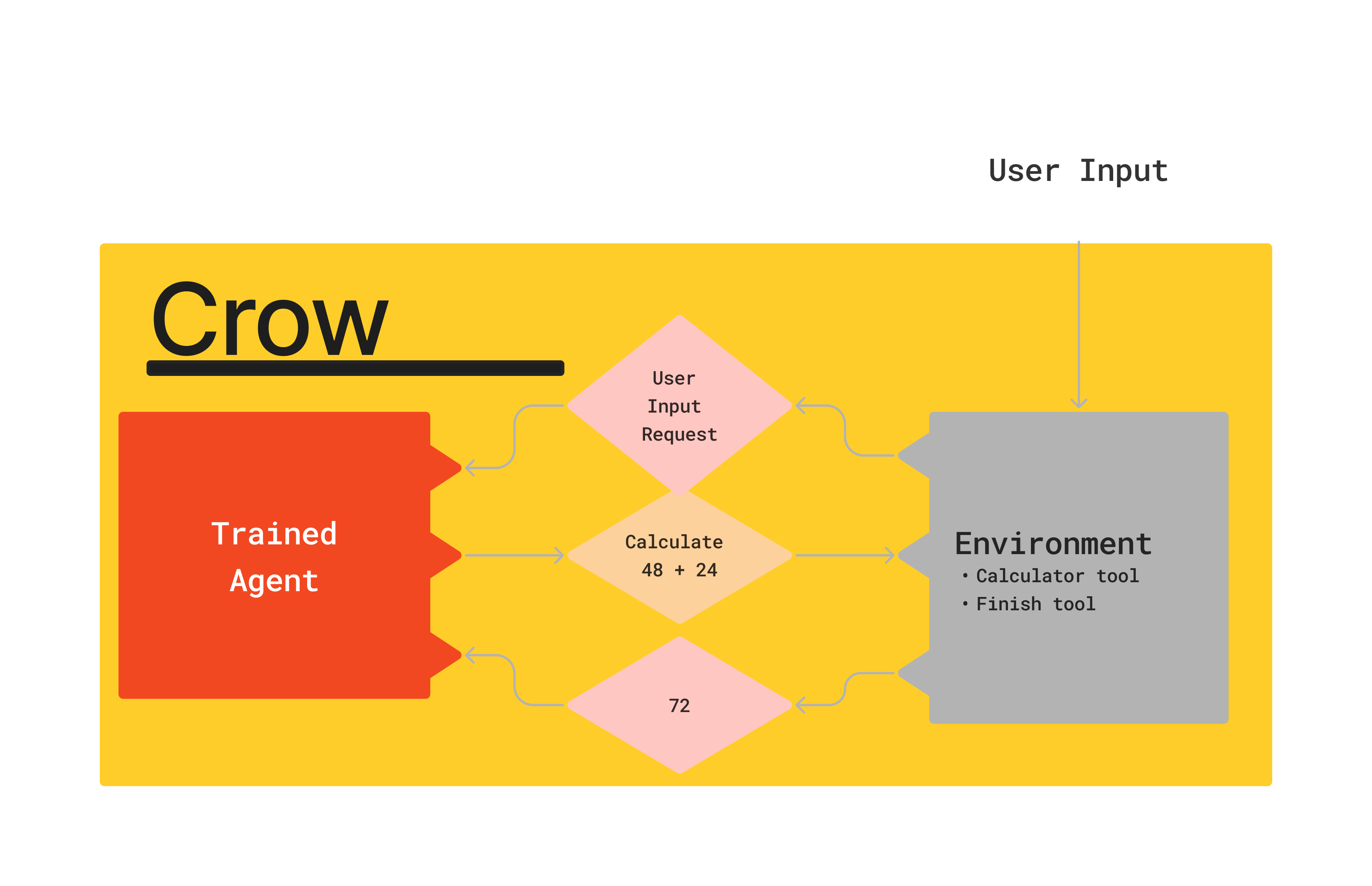
Crows
| Environment | Key Tools | |
|---|---|---|
| PaperQA | Literature Research | Search, Citation Traversal |
| ProteinCrow | Designing novel proteins | AlphaFold2, Molecular Dynamics |
| ChemCrow | Designing new molecules | Retrosynthesis, self-driving robotic lab |
PaperQA: an agent for literature research
Language agents achieve superhuman synthesis of scientific knowledge
Michael D. Skarlinski, Sam Cox, Jon M. Laurent, James D. Braza, Michaela Hinks, Michael J. Hammerling, Manvitha Ponnapati, Samuel G. Rodriques, Andrew D. White arXiv:2409.13740, 2024
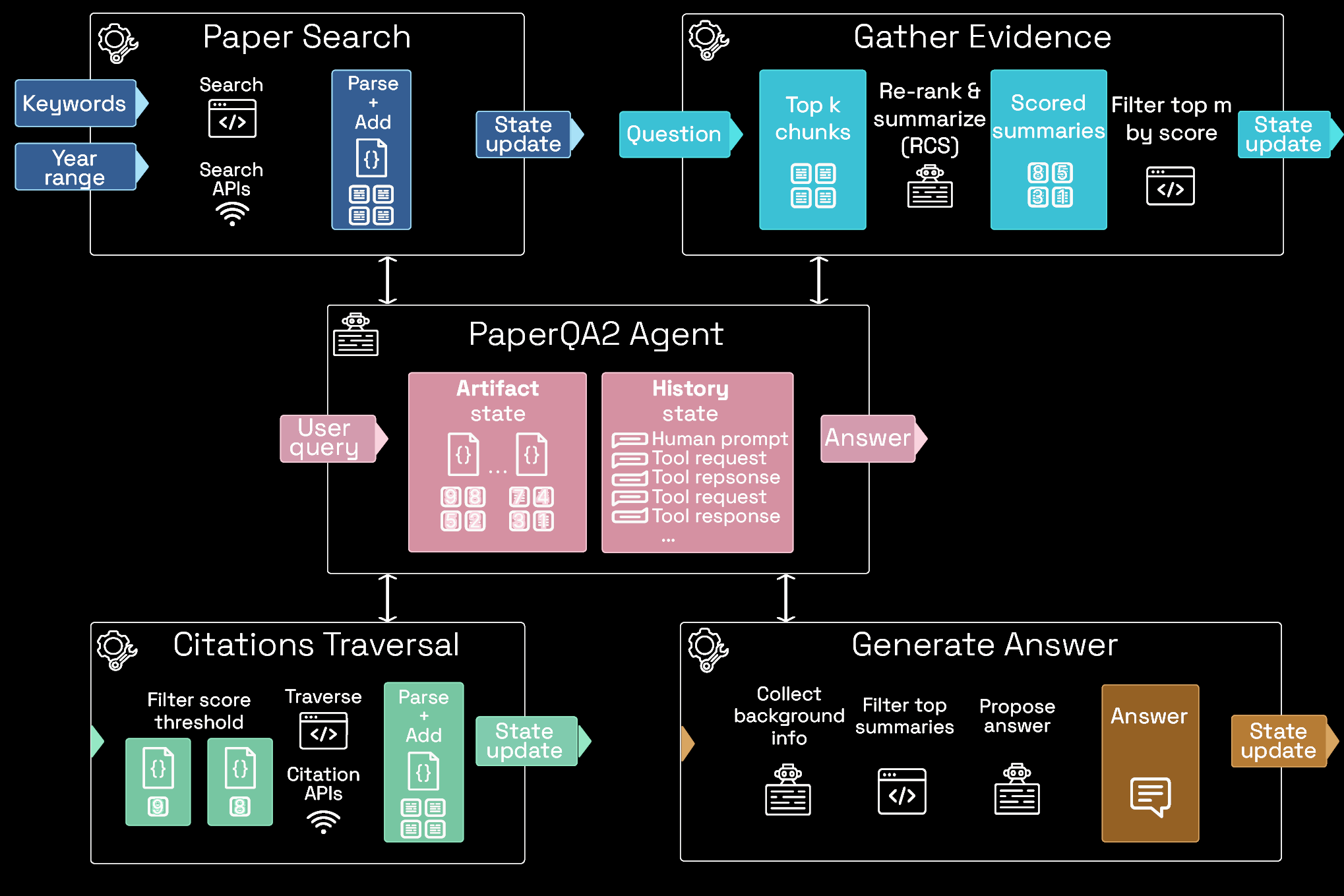
Better at answering questions than PhD biology experts
Accuracy vs Precision
Better than human written Wikipedia articles
Difference between PaperQA and Wikipedia
| WikiCrow | Wikipedia | |
|---|---|---|
| Unsupported Breakout (total) | 23 | 42 |
| Reasoning Issues | 12 | 26 |
| Attribution Issues | 10 | 16 |
| Trivial Statements | 1 | 0 |
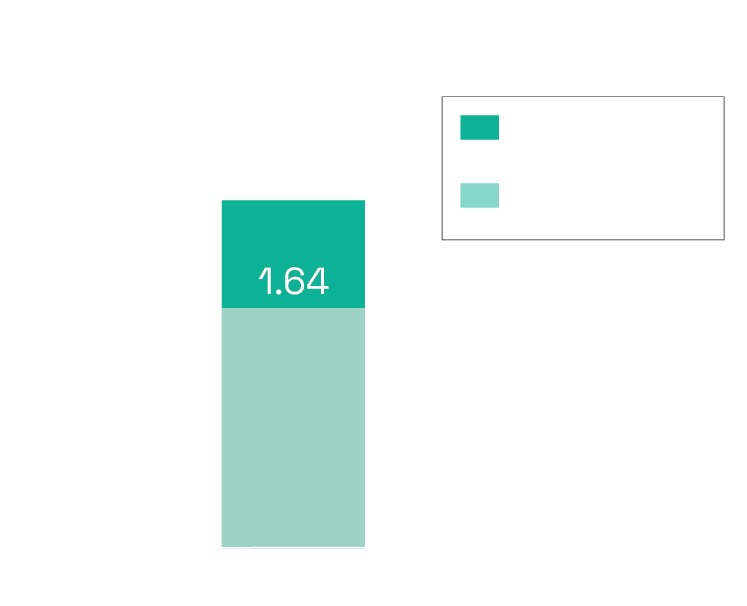
Can detect if a claim is contradicted anywhere in literature
Applications
WikiCrow
- Wikipedia articles for all 19,255 protein-coding genes
- Succeeded on 17,269
- Wikipedia had 3,639, so gain of 13,630
- 48 Hours
- Tasks per minute: 25
- Wiki page for all diseases every 3.5 days
- All arxiv papers per week 25,000 papers / month
- Check for contradictions (10x) 6.3M papers / year
- All Wikipedia (10x) every 3 weeks
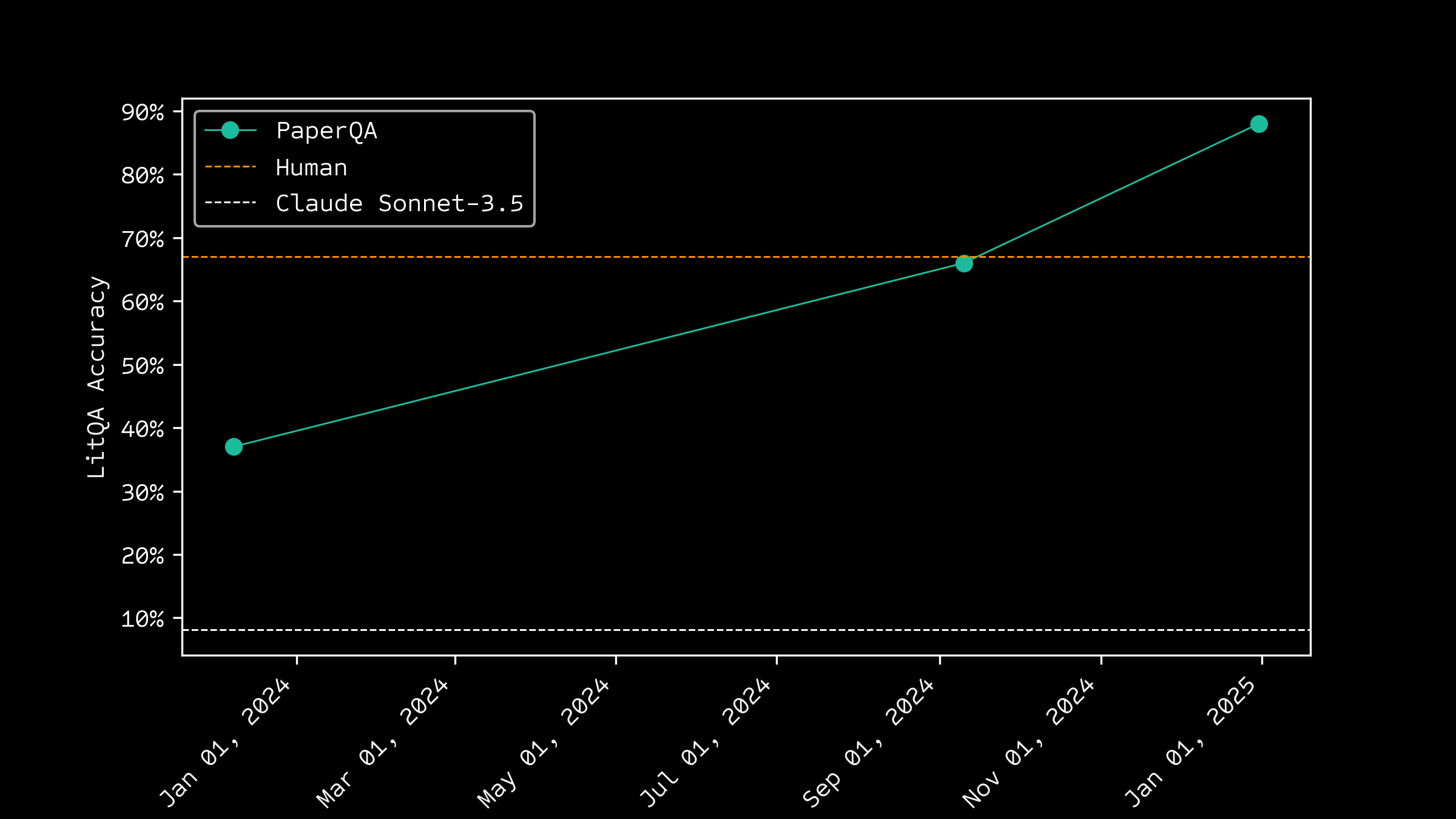
Progress on Accuracy